Alessandro Di Domizio
Founder of SPILLOproject / Algorithm and software developer
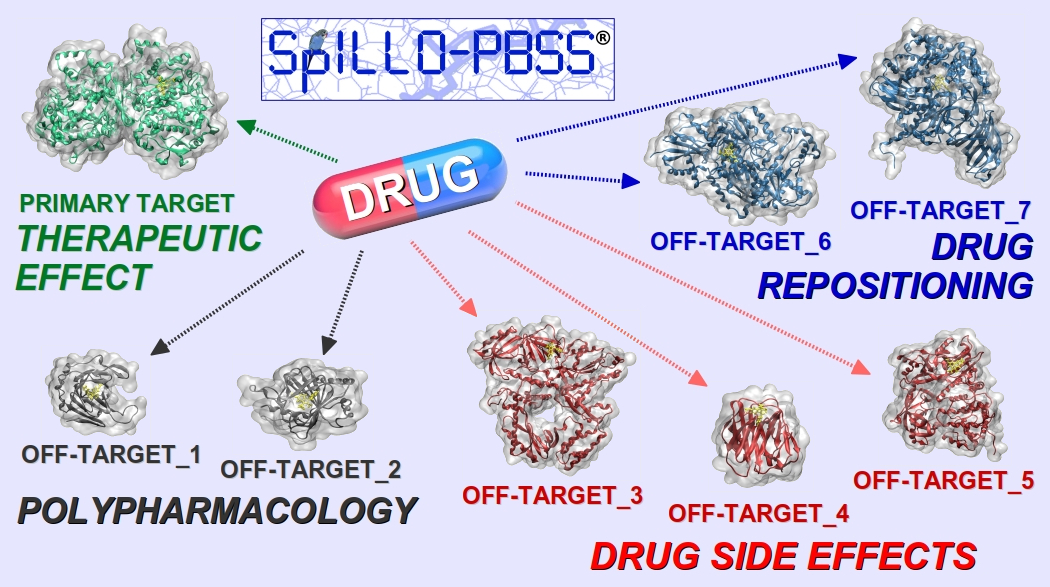
The property of a drug to act with multiple molecular targets usually represents a major drawback both in the Drug Discovery and Development process, where toxicity is responsible for the high failure rate (‘Attrition rate problem’) of drug candidates, and in medicine, where therapeutic agents often lead to side effects that can cause dose limitation or even discontinuation of treatment.
The innovative software SPILLO-Potential Binding Sites Searcher (SPILLO-PBSS [1], [2]), will be presented. It is designed to identify the biomolecular origin of the adverse and/or therapeutic effects of small molecules by a fast identification of their target and off-target proteins on a proteome-wide scale (e.g., the whole available human structural proteome (i.e., 15000+ protein 3D structures)).
The uniqueness of SPILLO-PBSS lies in its ability to find the potential binding sites (PBSs) on protein 3D-structures even if they are highly distorted and/or apparently inaccessible (e.g., closed, buried, occupied) to the small molecule, in comparison with an ideal conformation suitable for the binding.
Therefore, compared to traditional software, SPILLO-PBSS is more likely to find target and off-target proteins responsible for side-effects and/or for (additional) positive effects of the small molecule (e.g., main target identification, polypharmacology, drug rescue/repositioning).
The possibility of generating a clearer map of the drug-target interactions has remarkable implications both in the Drug Discovery and Development process and in Personalized Medicine, where treatments with the lowest toxic impact may be selected taking into account individual protein level differences.
According to the "bring your own problems" concept, the participants will be given the opportunity of evaluating (feasibility studies) the use of SPILLO-PBSS to solve their own specific research problems.
REFERENCE:
1) A. Di Domizio et al (2014). SPILLO-PBSS: Detecting hidden binding sites within protein 3D-structures through a flexible structure-based approach. J. Comp. Chem. 35 (27): 2005-2017.
2) SPILLO-PBSS Website: www.spilloproject.com
PRESENTED IN:
