Paolo Tosco
CRESSET
A macromolecular target interacts with a ligand by means of steric and electrostatic interactions. This means that, no matter which the chemical structure of a ligand is, if the steric and electrostatic field properties in its bioactive pose are complementary to the protein's active site, the ligand will have a high binding affinity.
Characterizing ligands by their molecular interaction fields, rather than by their structure, enables finding new and diverse actives by looking at the field similarity to known actives. This inherently allows the possibility for scaffold hopping and biosisosteric replacement, as long as the field properties are overall preserved [1,2].
Similarly to pharmacophore-based approaches, field-based models can be successfully applied also to targets whose x-ray structure is not available, provided that the binding mode and site are consistent across the ligand series.
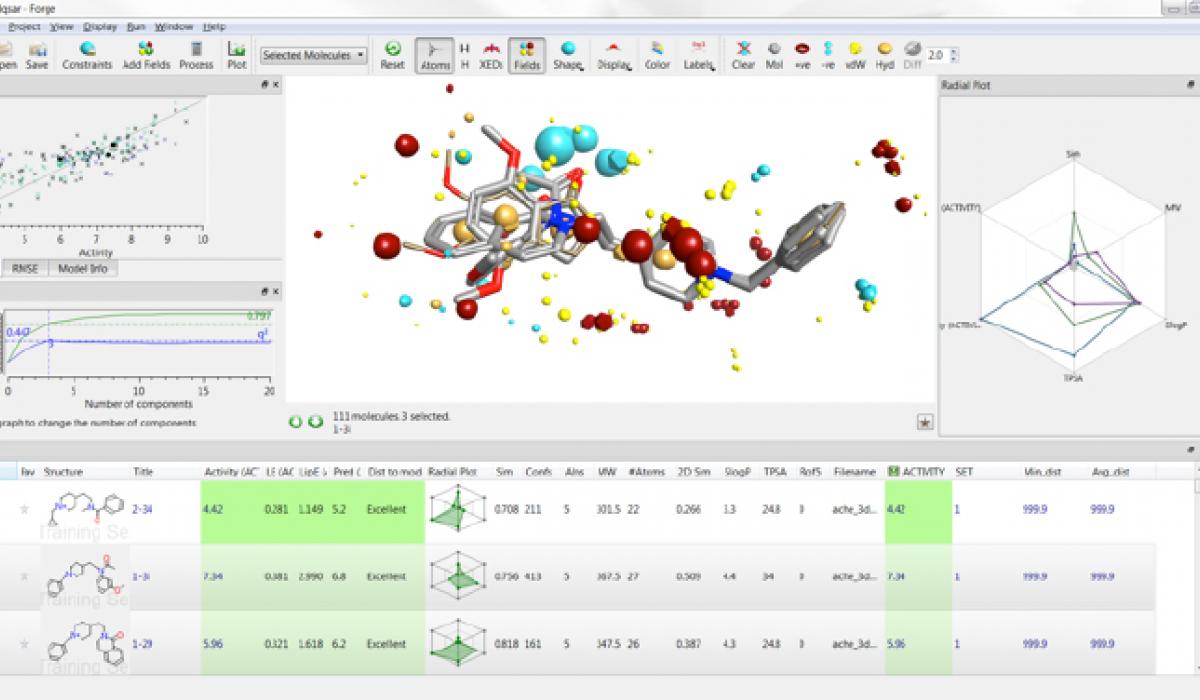
During this workshop Forge will be used to generate a binding mode hypothesis for a series of Plasmodium falciparum kinase inhibitors and to investigate structure-activity relationship across the series. The model obtained with Forge will be the starting point for a bioisosteric fragment replacement exercise using Spark in order to find new, structurally diverse, potential actives.
References
1. Cheeseright, T.; Mackey, M.; Rose, S.; Vinter, A. "Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation". J. Chem. Inf. Model. 2006, 46, 665-676.
2. Cheeseright, T. J.; Mackey, M. D.; Melville, J. L.; Vinter, J. G. "FieldScreen: Virtual Screening Using Molecular Fields. Application to the DUD Data Set". J. Chem. Inf. Model. 2008, 48, 2108-2117.
PRESENTED IN:
